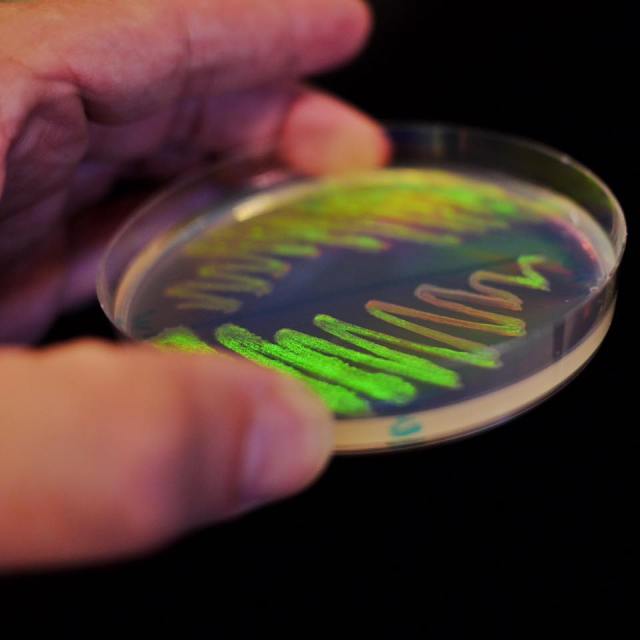
My time here at the Microbial Diversity Course at MBL has come to an end after visiting for the past 2 weeks with our lab’s MinION to sequence genomes from bacterial isolates that students collected from the Trunk River in Woods Hole, MA. (Photo by Jared Leadbetter of ‘Ectocooler’ Tenacibaculum sp. isolated by Rebecca Mickol).
In Titus Brown’s DIB lab, we’ve been pretty excited about the Oxford Nanopore Technologies MinION sequencer for several reasons:
1) It’s small and portable. This make the MinION a great teaching tool! You can take it to workshops. Students can collect samples, extract DNA, library prep, sequence, and learn to use assembly and annotation bioinformatics software tools within the period of 1 week.
2.) We’re interested in developing streaming software that’s compatible with the sequencing -> find what you want -> stop sequencing workflow.
3.) Long reads can be used in a similar way as PacBio data to resolve genome and transcriptome (!) assemblies from existing Illumina data.
Working with any new technology, especially from a new company, requires troubleshooting. While Twitter posts are cool, they tend to make it seem very easy. There is a MAP community for MinION users, but a login is required and public searching is not possible. In comparison to Illumina sequencing, there is not that much experience out there yet.
Acknowledgements
These are usually saved for the end, but since this is a long blog post, thought I would front-load on the gratitude.
I have really benefitted from blog posts from Keith Robison and lonelyjoeparker. Nick Loman and Josh Quick’s experiences have also been beneficial.
There is no match, though, for having people in person to talk to about technical challenges. Megan Dennis and Maika Malig at UC Davis have provided amazing supportive guidance for us in the past few months with lab space and sharing their own experiences with the MinION. I’m very grateful to be at UC Davis and working with Megan.
This trip was made possible by support from my PI, Titus Brown, who provided funding for my trip and all the flowcells and reagents for the MinION sequencing. It was necessary to have this 2 week block of time to focus on nothing else but getting the MinION to work, ask questions, and figure out what works (and what doesn’t).
Special thanks to Rebecca Mickol and Kirsten Grond in the Microbial Diversity course for isolating and culturing the super cool bacterial samples. Scott Dawson at UC Davis (faculty at the Microbial Diversity course) was instrumental in helping with DNA extractions. Jessica Mizzi assisted with library prep protocol development and troubleshooting. Harriet Alexander assisted with the assembly, library prep and showing me around Woods Hole, which is a lovely place to visit. Thank you also to the MBL Microbial Diversity Course, Hilary Morrison and the Bay Paul Center for hosting lab space for this work to take place.
https://www.flickr.com/photos/lpcohen/28192995104/
Files: https://github.com/ljcohen/dib_ONP_MinION
Presentation slides: https://docs.google.com/presentation/d/1Zqd1ayumdZqYc5e8bfeul8-57trKGwGdOFUdPc2-mIU/edit?usp=sharing
Immediately following the Woods Hole visit at MBL, I went to the MSU Kellogg Biological Station as a TA for NGS 2016 course and wrote a tutorial for analyzing ONP data:
http://angus.readthedocs.io/en/2016/analyzing_nanopore_data.html
Purchasing and Shipping
Advice: allow 2-3 months for ordering. We ordered one month in advance. While ONP customer service probably worked overtime to send our flowcells and Mk1B after several emails, chats, and calling in special favors, in the future it is unclear whether we can count on a scheduled delivery with students. Communication required ~dozen emails and we could never get confirmation that flowcells would arrive in time for the course. It turns out that our order had been shipped and arrived on time, however we did not know about it because a tracking number was not sent to us. It took about a day of emailing and waiting to track the boxes down. Thankfully, the boxes were stored properly in the MBL shipping warehouse.
Communicate with ONP constantly. Stay on top of shipments, ask for tracking numbers and confirmation of shipment. Find out where the shipment is being delivered, as the address you’ve entered may not be the one on the shipping box and your order will be delivered to the wrong place.
Flowcells
QC the flowcells immediately. Bubbles are bad:
https://www.flickr.com/photos/lpcohen/27996712883/in/dateposted-friend/
We ordered 7 flowcells (5 + 2 that came with the starter pack).The flow cells seemed to have inconsistent pore numbers and some arrived with bubbles. One flow cell had zero pores. They sent us a replacement for this flowcell within days, which was very helpful. However, for the flowcells that had bubbles, I was given instructions by ONP technical staff to draw back a small 15 ul vol of fluid to try to remove the bubble, then QC again. This did not work. The performance of these flowcells did not meet our expectations.
In communicating with the company, we were told that there was no warranty on the flowcells.
DNA Extractions
The ONP protocol says that at least 500-1000 ng clean DNA is required for successful library prep. Try to aim for more than this. Try to get as much DNA, as high molecular weight as possible. Be careful with your DNA. Do not mix liquids by pipetting. For the bacterial isolates from liquid culture, Scott Dawson recommended using Qiagen size exclusion columns to purify, and this worked really well for us. We started with ~2000 ug and used the FFPE repair step.
The ONP protocol includes shearing with the Covaris gtube to 8kb. When I eliminated this step to preserve longer strands, there was little to no yield and samples with adequate yield had poor sequencing results. In communicating with ONP about this, we suspected that the strands were shearing on their own somewhere during the multiple reactions, then either getting washed away during the bead cleanup steps, or the tether and hairpin adapters were sheared off so the strands were not being recognized by the pores.
We sequenced all three sets of DNA below (ladder 1-10kb). The Maxwell prep (gel below on the left) had a decent library quantity but the sequencing read lengths were not as long as we would have liked, which makes sense given the small smeary bands seen. (poretools stats report)

Library prep
When we first started troubleshooting the MinION, the protocols available through the MAP were difficult to follow in the lab. We needed a sheet to just print out and follow from the bench, so we created this:
https://docs.google.com/document/d/1EvxAyJFRu96_caWBEpCcQc7rfRV_Zm1LudScZA8lWCU/edit?usp=sharing
A few months ago, ONP came out with a pdf checklist for library prep, which is great:
Click to access ONP_MinION_lib_prep_protocol_SQK-NSK007.pdf
The library prep is pretty straight forward. One important thing I learned about the NEB Blunt/TA Master Mix:
Library prep and loading samples onto the flowcell can be tricky and nerve wracking for those who are not comfortable with lab work. I have >4 yrs of molecular lab experience knowing how to treat reagents, quick spins, pipetting small volumes, how to be careful not to waste reagents. One important point to convey to those who do not do molecular lab work often, is the viscous, sticky enzyme mixes that come in glycerol. You think you’re sucking up a certain volume, but an equal amount is often stuck to the outside of your pipette tip. You have to wipe it on the side of the tube to get it off so you don’t add this to your rxn volume, changing the optimal concentration and (probably the most important) also wasting reagent.
Other misc. advice:
- The calculation: M1V1 = M2V2 is your friend.
- Don’t mix by pipetting.
- Instead, tap or flick the tube with care.
- Quick spin your tubes often to ensure liquid is collected down at the bottom.
- Bead cleanups require patience and care while pipetting.
- Be really organized with your tubes (since there are a handful of reagent tubes that all look the same). Use a checklist and cross off each time you have added a reagent.
These are the things I take for granted when I’m doing lab work on a regular basis. It takes a while to remember when I’m in the lab again after taking a hiatus to work on computationally-focused projects.
https://www.flickr.com/photos/lpcohen/28040963554/in/dateposted-friend/
Computer Hardware
In October 2015 last year when we were ordering everything to get set up, the computer hardware requirements for the MinION were: 8GB RAM and 128 SSD harddrive with i7 CPU. This is what we ended up ordering (which took several weeks to special order from the UC Davis computer tech center):
DH Part#: F1M35UT Manufacturer: HP Mfr #: F1M35UT#ABA HP ZBook 15 G2 15.6″ LED Mobile Workstation Intel Core i7 i74810MQ Quadcore (4 Core) 2.80 GHz 8 GB DDR3L SDRAM RAM 256 GB SSD DVDWriter NVIDIA Quadro K1100M 2 GB Windows 7 Professional 64bit (English) upgradable to Windows 8.1 Pro 1920 x 1080 16:9 Display Bluetooth English Keyboard Wireless LAN Webcam 4 x Total USB Ports 3 x USB 3.0 Ports Network (RJ45) Headphone/Microphone Combo Port
- Windows sucks
- There’s a new GUI (graphical user interface) for MinKnow in the past months. It’s annoying to get used to this, but in general not too bad.
- The poretools software to convert .fast5 to .fastq is buggy on Windows and does not play well with MinKnow. There’s probably a way to get them both to work, but I’ve already spent ~2-4 hrs of troubleshooting this issue, so am done with this for now. Instead, we’ve been uploading .fast5 to a Linux server, then running poretools on there.
- MinKnow python scripts crash sometimes during the run! You can open the MinKnow software again, start the script again, and it should start the run from where it left off.
- Use the 48 hr MinKnow script for sequencing.
- Our flow of data goes from raw signal from the MinION (laptop) -> upload to Metrichor server for basecalling -> download to external hard-drive (“pass” or “fail” depending on the Metrichor workflow chosen, e.g. 1D or 2D or barcoding) -> plug external hard-drive to Linux or Linux laptop (for some reason this is easier on Linux laptop rather than Windows…) for transfer to Linux server -> on the Linux server, run poretools software to convert to fastq/fasta -> analysis
- This all seems kind of ridiculous. If there is a better way, please let us know!
Workshops
In a future workshop setting, where students are doing this for the first time but we have more experience now, a potential schedule could go something like this:
Day 1: Collect sample, culture
Day 2: Extract DNA, run on gel, quantify
Day 3: Library prep, sequence (this will be a long day)
Day 4: Get sequences, upload, assess reads, start assembly
Day 5: Evaluate assembly, Annotate
This is similar to the schedule arranged for Pore Camp, run by Nick Loman at the University of Birmingham in the UK. They have some great materials and experiences to share:
Cost
- Still unknown what the cost is per sample.
- Cost of troubleshooting?
I’ve put together a quick ONP MinION purchasing sheet:
https://docs.google.com/spreadsheets/d/1yBncz75kgwExCXy7sC9LsMaDGs8OJJJGg9f4o3DcoQE/edit?usp=sharing
Generally, these are the items to purchase:
- Mk1B starter pack came with 2 flowcells
- computer
- ONP reagents
- third-party reagents (NEB)
Getting Help
- MAP community has some answers
- There is no phone number to call ONP. In contrast, Illumina has a fantastic customer service phone line, with well-trained technicians on the other end to answer emergency phone calls. Reagents and flowcells are expensive. When you’re in the lab and there is a problem, like a bubble on the flowcell or a low pore number after QC, it is often necessary to call and talk to a person on the phone to ask question so you don’t waste time or money.
- I’ve had many good email conversations with ONP tech support, but there is no substitute to calling someone on the phone and discussing a problem. Often, there are things to work on after the email and it is difficult to follow up by going back and forth with email.
- LiveChatting feature on the ONP website is great! (During UK business hours, there is a feature at the bottom of the store website that says “Do you have a question?”. During off hours it says “Leave a message”.
I realized through this process that I had lots questions and few answers. The MAP has lots of forum questions but few manuals. Phrase searching sucks. If you search for a phrase in quotes, it will still search for individual words. For example:
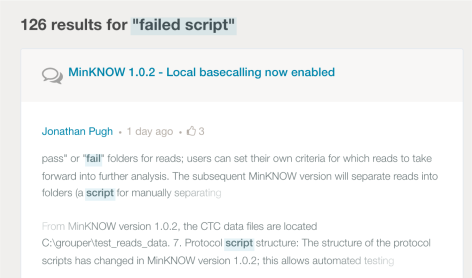
Remaining Questions:
1. Why does the number of flow cell pores fluctuate? What is the optimal pore number for a flow cell?
2. What is the effect of 1D reads on the assembly? Can we use the “failed” reads for anything?
3. How long will a run take?
4. How much hard-disk space is required for one run?
5. When are the reads “passing” and when are they “failing”? Is there value to the failing reads?
6. How can we get the most out of the flow cells? There seem to be a lot of unknowns related to the efficiency of the flowcells. We tried re-using a washed flow cell. There were >400 pores in the flow cell during the initial QC. After we loaded the library and started the run, the pore numbers were in the 80s-100s. 2 hrs later, this number dropped down to ~30s. I added more library, and the pore numbers never increased again. Is this a result of the pore quality degrading? The next morning, loaded more library again. Not much change. Decided to switch flowcells and try a new one.
7. Are there batch effects of library prep and/or flowcells? Should we be wary of combining reads from multiple flowcells?
Future
In the future, the aim is to move away worrying about the technology details and focus on the data analysis and what the data mean. The goal should be to focus on the biology and why we’re interested in sequencing anything and everything. What can we do with all of this information, now that we can sequence a genome of a new bacterial species in a week?
Feel free to comment and contact!


Hi! Great post. You obviously did better than my 1 usable read from 3 flowcells (which I blogged on recently)!
Just a few notes on items you mentioned that have changed recently. Local base calling is now available, so that slightly simplifies things. Isn’t there a Mac MinKNOW? That would also simplify. If you get WinSCP (free), then you can move files over the net – I’m nbad about peeking at the data as it comes in.
ONT now has a warranty on number of pores at Platform QC – but you need 6o test within 5 days of receipt.
The buzz on fail reads has been that they don’t have usable data. Assembling from 1D is a very interesting area to explore with the Rapid 1D kit now available – and should work since the data quality should be PacBio–ish.
I hope ONT takes your suggestions on support & the search interface seriously – finding the right info is too often a chore
Great questions at end – other than perhaps MARC, I don’t believe anyone has looked at reproducibility, and the idea of batch effects certainly deserves attention.
Thanks for your comment and your blog!
– Will look into WinSCP, thank you.
– Yes, MinKNOW 0.51.3 onwards will run on MAC OS X: https://nanoporetech.com/sites/default/files/s3/2016-07/Computer%20requirements%20v3%20Jul2016.pdf
When we started acquiring stuff in 2015, Windows OS w/ SSD and i7 processor was the only option so that’s what we have.
– Yep, we did QC on flowcells upon receipt. A week later, QC again with some similar pore numbers but some lower. After this post we ended up complaining again about one flowcell in particular with >900 pores, then >600 a week later, then 435 pores a few days later when we actually did the run (after being told by ONT to use it). The run only had a few thousand reads, so they sent a replacement.
– We were told that the ‘starter pack’ flowcells did not have a warranty.
– Have plans to look more in-depth with 1D reads. Do you know how the hairpin is dealt with if analyzing 1D reads from 2D prep?
Pingback: Links 8/24/16 | Mike the Mad Biologist
Thank you for posting details about the process and troubleshooting, I’m sure this will come in very handy as we begin using our own MinION. I’m curious too about the fluctuating pore number, would be useful to know how/why this changes (including instances where it increases).
Extremely useful post to read before getting my hands dirty with my first MinION starter pack. My expectations are dampened, perhaps appropriately. I can second your concerns about the “customer service” side of the Oxford Nanopore system: both the community search tool and the documentation remain less than optimal. Even prior to getting my first shipment I am in the dark about how MinKNOW is supposed to work and how to engineer my pipeline (particularly if I want to do anything approaching a live analysis).
Any additional experience since this post?
Thanks for your blog post. I’m a Masters student at the Natural History Museum in London and during my time here I will hopefully be using the MinION to do some RNA seq experiments using R9.4 flow cells. Reading this has made me feel better about what we can and can’t expect from the MinION in terms of throughput and user friendliness. From my very limited understanding on the MinION there appears to be some basic questions from the scientific community that are still unanswered on the ONT community site like what the expected lifetime of a flow cell is and expected output from each flow cell. If anyone is aware of a place where these questions have been answered a reply would be appreciated.
Excelent post! Really help
Pingback: Q&A at Porecamp TAMU, June 2017: From the Ebola Outbreak to Sequencing on Mars | Lisa Johnson
Hi, It is such an excellent article, thanks for sharing this article with us.Also for further informations visit https://hard-drives.co/wd-external-hard-drive-not-recognized-windows-10